ENGENISTM siRNA Test System Cat # N0070 £580 manual (pdf) vector sequences
Luciferase-based ENGENISTM (Entire Gene/cDNA or Isolated Sequence) siRNA Test System is designed to provide a quantitative approach for evaluation of a potential of any DNA sequence to serve as an efficient target for RNA interference (RNAi). Current computer-based RNAi target searching algorithms are not perfect, leaving the probability of selected sequence to be a good target from 30% to 60%. It is therefore clear that the ability of computer-selected sequence to induce RNA interference has to be confirmed experimentally. The ENGENISTM siRNA Test System allows for screening a large number of potential siRNA target sequences in simple transient transfection experiments, measuring the reduction in reporter (firefly Luciferase from Photinus pyralis) gene expression.
In most commercially available vectors and kits intended for the same purpose, the gene silencing potential of a target sequence is tested on the fused transcript bearing both reporter gene and gene of interest. In such systems some unpredictable factors like chimeric mRNA folding and target accessibility may affect the observation of RNAi. Also the distance between the target site and reporter gene coding region along the mutual messenger RNA plays a role in the reporter gene silencing: even if mRNA is cleaved at the point of siRNA target, translation of the reporter gene can still go on till the RNA degradation by nonspecific nucleases reaches the reporter gene coding region. Such kind of effects may obscure the real efficiency of particular targets.
Unlike other kits, the ENGENISTM siRNA Test System offers two possibilities for evaluation of the target sequence efficiency: on the one hand, using entire gene technology, and on the other hand, using a technology with isolated target sequences per se. In the latter case, short target sequences are located at defined positions immediately upstream or downstream of the Luciferase gene coding region. For entire gene technology, two variants of transcript can be generated by placing the gene of interest either in front of Luciferase coding region or behind it.
Independent evaluation of the reporter gene silencing corresponding to two different positions of isolated target sequence or entire target gene permits to avoid or reduce the influence of target accessibility factor and other factors related to RNA conformation, thus making comparison between various targets more reliable. Another advantage of the ENGENISTM siRNA Test System is that it makes possible evaluation of the siRNA target potential even in those cases when the gene of interest is not available as full-size cDNA clone.
The ENGENISTM siRNA Test System consists mainly of four vectors: one effector plasmid (psiRNA) producing double-stranded siRNA, and three reporter plasmids (psiTEST-target-LUC, psiTEST-LUC-target, psiTEST-target-IRES-LUC) expressing Luciferase reporter gene fused to siRNA target sequence or the target gene cDNA.
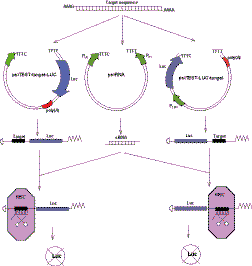
siRNA testing on isolated targets (Fig. 1) is based on the fact that the nucleotide sequence of siRNA is identical to the sequence of corresponding siRNA target. Thus the same short synthetic DNA fragment has to be cloned into three linearized plasmids to generate siRNA coding sequence in effector plasmid psiRNA and siRNA target sequence in two reporter plasmids psiTEST-target-LUC and psiTEST-LUC-target.
Fig. 1. Construction of effector and reporter plasmids for siRNA testing on isolated targets and their mechanism of action
In the frames of entire gene technology, the ENGENISTM siRNA Test System provides two supercoiled vectors psiTEST-LUC-target and psiTEST-target-IRES-LUC containing multiple cloning sites (MCS) intended for target gene cloning and subsequent generation of a fused transcript (Fig. 2). Encephalomyocarditis virus Internal Ribosomal Binding Site (IRES) of the reporter plasmid psiTEST-target-IRES-LUC enables Luciferase translation from the second cistron of the fused transcript. MCS structures and vector backbones are presented in the Section VI.
Fig. 2. Construction of effector and reporter plasmids for siRNA target testing on fused mRNA and their mechanism of action.
siRNA expression from the effector plasmid psiRNA is driven by dual promoter expression cassette bearing human U6 and H1 small nuclear RNA promoters in opposite orientation to each other. Such dual promoter constructs were shown to efficiently express double-stranded siRNA molecules (1-2) which could be directly accepted by RNA Interference Searching Complex (RISC), the multi-enzyme complex with RNase activity specifically digesting messenger RNA at the siRNA target site (3). This permits to avoid the stage of dicer treatment which is necessary for hairpin siRNA producing vectors to convert single-stranded hairpin shRNA to double-stranded siRNA.
Detailed structure of the double-promoter siRNA expressing cassette is shown in Fig. 3. siRNA coding sequence N1-N19 is placed between the U6 and H1 promoters. RNA synthesis driven by the U6 promoter starts from the nucleotide N1 which has to be G, and terminates at the stretch of 5 T invading the body of H1 promoter. As a result, produced RNA has 2 or 3 uridines at its 3' terminus. Similarly, the RNA produced from the H1 promoter starts with the nucleotide N'19, terminates at 5 T stretch at the beginning of the opposite U6 promoter, and also bears 2 or 3 uridines at its 3' terminus. Two halves of siRNA join together, forming the functional double-stranded siRNA with protruding 3'-termini.
A mixture of siRNA-producing effector plasmid and one of the target-bearing reporter plasmids has to be cotransfected transiently into mammalian cells. Measuring the reporter gene silencing due to the effect of RNA interference will give a quantitative evaluation of the siRNA target potential.
Fig. 3. siRNA expression from the dual promoter effector plasmid psiRNA.